
Sleep, biological rhythms, and infection-induced neuroinflammation
In the brain, I discovered a subset of B cells, termed regulatory B cells with anti-inflammatory and protective functions that could limit parasite replication in this tissue (3). Intriguingly, I also found autoreactive B cells that produced antibodies against a wide range of host antigens in both mice and humans, highlighting that sleeping sickness is an autoimmune disorder affecting the brain (4).
These results highlight the dual role that B cells play in the central nervous system during chronic challenges such as those resulting from sleeping sickness. We are now exploring i) the origins of the B cells identified in the brain during infection, ii) the mechanisms by which they control neuroinflammation and neurodegeneration, and iii) how these B cells control circadian rhythms and sleep and architecture.
In collaboration with Dr. Andras Bikov (NHS England) we are also immunophenotyping patients with various sleep disorders, including narcolepsy, idiopathic hypersomnias parasomnias, and sleep apnoea. Our vision is that both our pre-clinical and clinical work will provide unparalleled insights into the immunological mechanisms underlying complex sleep disorders
A)

B)

C)

D)

A) Predicted localisation of brain-dwelling B cells in the healthy and chronically infected murine brain using spatial transcriptomics, which enables us to generate hypothesis regarding tissue localisation of cell types of interest in an unbiased manner. B) Example of running wheel activity, using to monitor circadian behaviour, from naive and infected animals. The dark-shade bars at the top represent the active period (night time) whereas the light-shaded boxes represented the rest period (day time). C) Jessica Costa conducting surgeries to implant the subcutaneous telemetry implants that we use to monitor electrocorticogram (ECoG) and electromyogram (EMG) that are necessary to measure sleep architecture. D) Representative ECoG and EMG from a healthy mouse, revealing various sleep and wakefulness stages.
Host-pathogen interactions & Adipose tissue responses to T. brucei infection (Sinton lab)
African trypanosomes establish complex interactions with the immune system, enabling them to sustain chronic infections in the host. In the past, we applied several cutting-edge methods, including single cell and spatially transcriptomics, to study host-trypanosome interactions in murine models of infection. In the skin, we discovered a novel communication crosstalk between skin-resident T cells and subcutaneous adipocytes mediated by IL-17 that balance local immune responses, restraining parasite replication in vivo (1,2).

As part of a Wellcome Trust Early Career Fellowship, Matthew Sinton (lab co-lead) is now building the foundations of his laboratory focussing on exploring how local tissue responses in the adipose tissue induces changes in feeding behaviour during infection-induced cachexia.
Pathogen biology & tool development to study host-pathogen interactions
In the past, we have used several molecular biology tools to identify the mode of action of drugs used in the field to treat sleeping sickness (. I have worked with physicists to implement whole brain clearing and imaging using optical mesoscopy, enabling imaging infected host tissues across a large tissue volume with subcellular resolution (6,7). This enabled us to identify discrete brain areas with exacerbated neuroinflammation in response to infection, indicating that the tissue responses are likely to be spatially heterogeneous. I also developed an in vitro system using human brain organoids to evaluate the responses of human tissue against the pathogen T. brucei gambiense, the main causing agent of sleeping sickness in humans (8). Moving forward, we will continue to apply and develop novel methods (e.g., sLP-mCherry to cell-cell interaction studies in vivo) to decipher the “cellular neighbourhood” surrounding parasites in tissues, and how those interactions between parasites and host define disease outcomes.
A)

B)

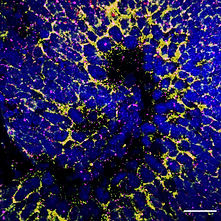
A) The human pathogen Trypanosoma brucei gambiense induces an upregulation of transcriptional signatures associated with neurogenesis and cell differentiation in iPSC-derived human brain cortical organoids in vitro. B) Representative images of in situ hybridisation for Gfap (yellow) and Neun (pink) from iPSC-derived human brain cortical organoids either left untreated (naive - left) or exposed to. T. b. gambiense for 72 hours (right). Scale bar, 50 micrometers.
Collaborators
We have ongoing collaborations with several groups both at the University of Manchester and elsewhere
University of Manchester:
University of Oxford:
Stanford University:
University of Oslo:
University of Belo Horizonte (Brazil):
University of Bordeaux:
The lab is generously funded by the following funding bodies:
Funding
Wellcome Trust Career Development Award
(2025 - 20233)



AMS Springboard Award
(2024 - 2026)
Research grant
(2024 - 2025)

